In this blog post, we will download the V-DEM datasets with their vdemdata package. It is still in development, so we will use the install_github() function from the devtools package
devtools::install_github("vdeminstitute/vdemdata")
library(vdemdata)And really quickly we can download the dataset with one line of code
vdemdata::vdem -> vdemWe can use the find_var function to get information on variables based on keywords.
For example, we can look up variables that are concerned with protest mobilization.
vdemdata::find_var("mobilization") -> mob
mob %>% names
[1] "question_id" "question_number" "metasection"
[4] "name" "vartype" "cb_section"
[7] "tag" "projectmanager" "question"
[10] "clarification" "responses" "ordering"
[13] "scale" "answertype" "sources"
[16] "notes" "datarelease" "citation"
[19] "coverage" "subsetof" "crosscoder_aggregation"
[22] "aggregation" "ccp_tag" "clean_tag"
[25] "survey_id" "vignettes_used" "old_tag"
[28] "compiler" "clarification_historical" "codebook_id"
[31] "conthistmerge" "histmerged" "years"
[34] "hist_outside_coding" "additional_versions" "cleaning"
[37] "date_specific" "available_versions" "cont_outside_coding"
[40] "overlap_use_hist" "is_party" "cb_section_type"
[43] "defaultdate" "convergence" "cy_aggregation"
[46] "no_update"

Or download the entire codebook:
vdemdata::codebook -> vdem_codebookAnd we can look at information for a specific variable
vdemdata::var_info("e_regionpol_6C") -> region_info
region_info$responses
1: Eastern Europe and Central Asia (including Mongolia and German Democratic Republic)
2: Latin America and the Caribbean
3: The Middle East and North Africa (including Israel and Türkiye, excluding Cyprus)
4: Sub-Saharan Africa
5: Western Europe and North America (including Cyprus, Australia and New Zealand, but excluding German Democratic Republic)
6: Asia and Pacific (excluding Australia and New Zealand; see 5)"
For our analysis, we can focus on the years 1900 to 2022.
vdem %<>%
filter(year %in% c(1900:2022))And we will create a ggplot() object that also uses the Five Thirty Eight theme from the ggthemes package.
Click here to read more about the ggthemes options.

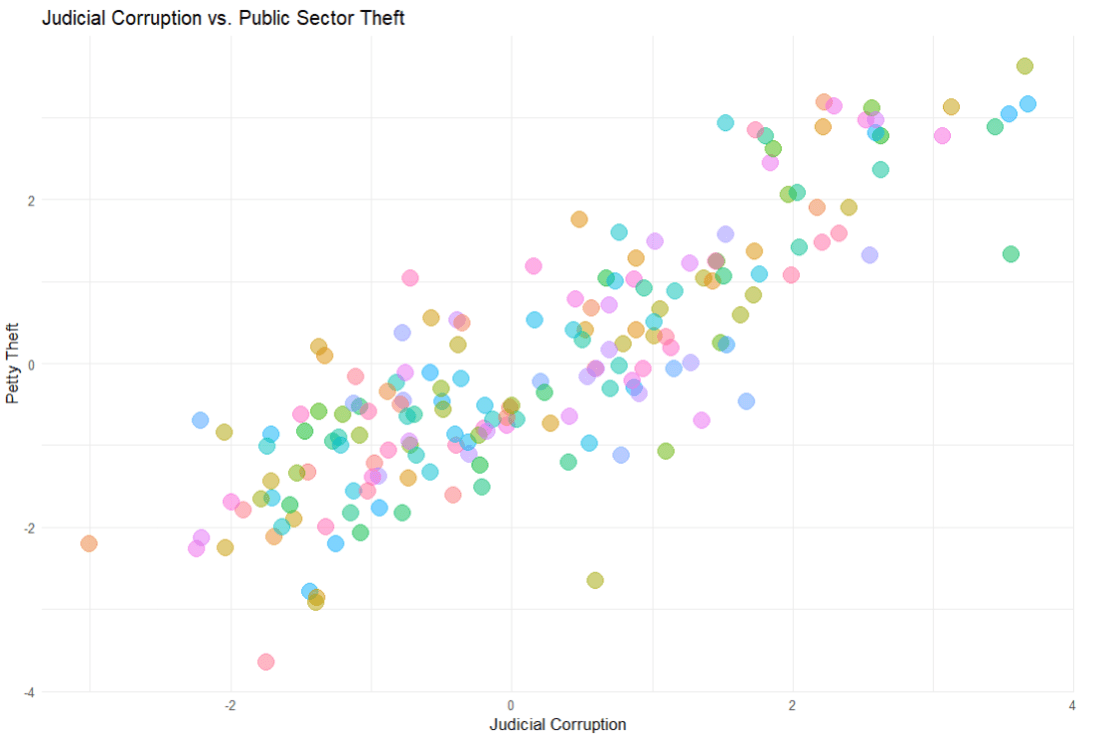
In the V-DEM package, we will look at a scatterplot of CSO consultation (v2cscnsult) and democracy score (v2x_polyarchy).
v2cscnsultasks are major civil society organizations (CSOs) routinely consulted by policymakers on policies relevant to their members?
v2x_polyarchyexamines to what extent is the ideal of electoral democracy in its fullest sense achieved?
First, find below the packages we will need to install and load
install.packages("gganimate")
install.packages("transformr") # sometimes needed as a dependency
library(gganimate)And we plot our graph:
my_graph <- ggplot(vdem, aes(x = v2cscnsult,
y = v2x_polyarchy,
group = year)) +
geom_point() +
ggthemes::theme_fivethirtyeight() +
theme(text = element_text(size = 12), # Default text size for all text elements
plot.title = element_text(size = 20, face="bold"),
axis.title = element_text(size = 16),
axis.text = element_text(size = 14),
legend.title = element_text(size = 14),
legend.text = element_text(size = 12)) In the themes argument, we can change the size of the text for the various parts of the ggplot (legends, axes etc.)
To make the ggplot object animated, we use the transition_time(year) function from the gganimate package.
Also we can add a subtitle the displays the year and time frame in the graph.
animated_graph <- my_graph +
transition_time(year) +
labs(title = "CSO consultation and Polyarchy Democracy",
subtitle = "Time: {frame_time}",
caption = "Source: VDEM 1900 to 2022",
x = "CSO Consultation",
y = "Polyarchy")And we can change how we render the graph with the animate() function.
We choose duration = 15 so that the gif lasts 15 seconds
We set frames per second to 20 fps (the higher the number, the smoother the gif changes, but the longer it takes to load)
And finally we can choose a special renderer that makes the gif more smooth too.
Finally we can save the gif to our computer (so I can upload it here on this blog)
animate(animated_plot, duration = 15, fps = 20, renderer = gifski_renderer()) -> CSO_poly_gif
anim_save("animated_plot.gif", animation = CSO_poly_gif)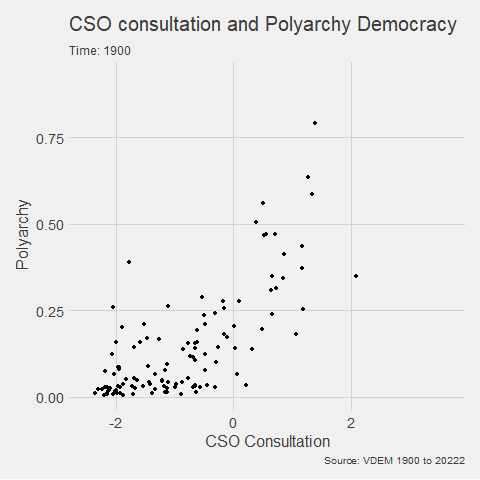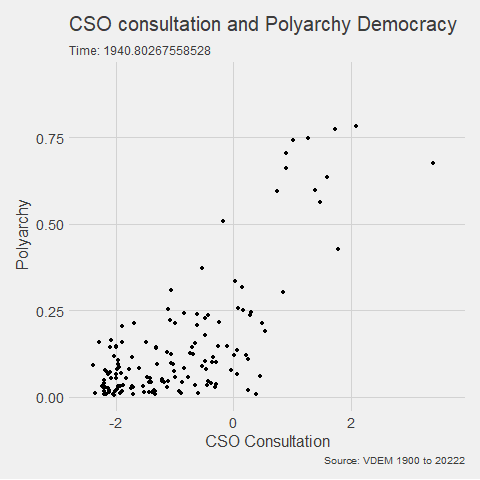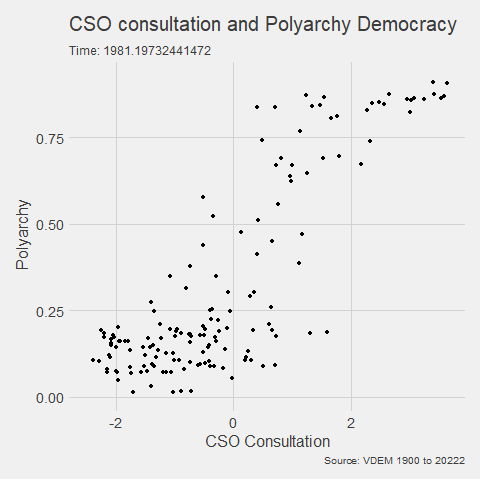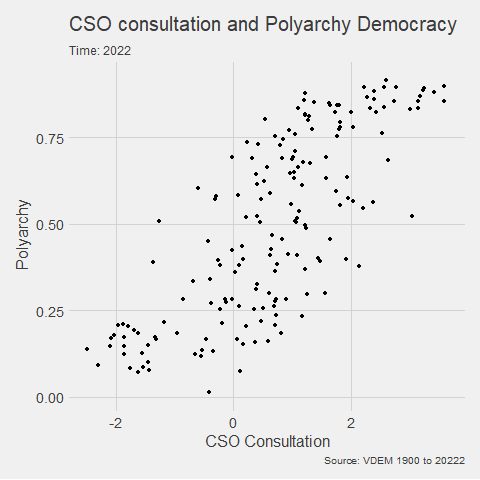
We can make a few changes so that it is divided by region and adds colors:
Notice the change to subtitle = "Year: {as.integer(frame_time)}" so it only uses the year, not the year and frame rate.
ggplot(vdem, aes(x = v2cscnsult,
y = v2x_polyarchy,
group = year,
size = e_pop,
colour = as.factor(e_regionpol_6C))) +
geom_point(alpha = 0.7, show.legend = FALSE) +
ggthemes::theme_fivethirtyeight() +
theme(text = element_text(size = 12),
plot.title = element_text(size = 20, face="bold"),
axis.title = element_text(size = 16),
axis.text = element_text(size = 14),
legend.title = element_text(size = 14),
legend.text = element_text(size = 12)) +
facet_wrap(~e_regionpol_6C) +
transition_time(year) +
labs(title = "CSO consultation and Polyarchy Democracy",
subtitle = "Year: {as.integer(frame_time)}",
caption = "Source: VDEM 1900 to 2022",
x = "CSO Consultation",
y = "Polyarchy") 
Next, we can animate a map
library(sf)
library(rnaturalearth)First we download a world object with the longitude and latitude data we need.
Click here to read more about the rnaturalearth package
my_map <- ne_countries(scale = "medium", returnclass = "sf")And we merge the two data.frames together
my_map %<>%
mutate(COWcode = countrycode::countrycode(sovereignt, "country.name", "cown"))
vdem_map <- left_join(vdem, my_map, by = c("COWcode"))Set up some colors for the map
colors <- c("#001427", "#708d81", "#f4d58d", "#bf0603", "#8d0801")
Next we draw the map:
vdem_map %>%
filter(year %in% c(1945:2023)) %>%
filter(sovereignt != "Antarctica") %>%
group_by(country_name, geometry) %>%
summarise(avg_polyarchy = mean(v2x_polyarchy, na.rm = TRUE)) %>%
ungroup() %>%
ggplot() +
geom_sf(aes(geometry = geometry, fill = avg_polyarchy),
position = "identity", color = "#212529", linewidth = 0.2, alpha = 0.85) +
geom_tile(data = data.frame(value = seq(0, 1, length.out = length(colors))),
aes(x = 1, y = value, fill = value),
show.legend = FALSE) +
scale_fill_gradientn(colors = colors,
breaks = scales::pretty_breaks(n = length(colors)),
labels = scales::number_format(accuracy = 1)) +
theme_minimal()
When I made the first attempt to animate the polyarchy democracy data on the map, it was a bit of an affront to the senses:

So attempt number two involved a bit more wrangling!
vdem_map %>%
filter(year %in% c(1945:2023)) %>%
filter(sovereignt != "Antarctica") %>%
ggplot() +
geom_sf(aes(geometry = geometry, fill = v2x_polyarchy, group = year),
position = "identity", color = "#212529", linewidth = 0.2, alpha = 0.85) +
theme_minimal() +
scale_fill_gradientn(colors = colors,
breaks = scales::pretty_breaks(n = length(colors)),
labels = scales::number_format(accuracy = 3)) +
transition_time(year) +
labs(title = "Polyarchy Democracy annual global changes",
subtitle = "Year: {as.integer(frame_time)}",
caption = "Source: VDEM 1900 to 2023",
x = " ",
y = " ",
fill = "Democracy Score") -> my_plot
animate(my_plot, duration = 40, fps = 40,
renderer = gifski_renderer()) -> map_gif
anim_save("animated_plot_3.gif", animation = map_gif)